Practical 2¶
In this practical we will start interacting more with Python, practicing on how to handle data, functions and methods. We will see several built-in data types and then dive deeper into the data type string.
Modules¶
Python modules are simply text files having the extension .py (e.g. exercise.py). When you were writing the code in the IDE in the previous practical, you were in fact implementing a module.
As said in the previous practical, once you implemented and saved the code of the module, you can execute it by typing
python3 exercise1.py
(which in Windows might be python exercise1.py, just make sure you are using python 3.x) or, in Visual Studio Code, by right clicking on the code panel and selecting Run Python File in Terminal.
A Module A can be loaded from another module B so that B can use the functions defined in A. Remember when we used the sqrt function? It is defined in the module math. To import it and use it we indeed wrote something like:
[1]:
import math
A = math.sqrt(4)
print(A)
2.0
Note
When importing modules we do not need to specify the extension “.py” of the file.
Objects¶
Python understands very well objects, and in fact everything is an object in Python.
Objects have properties (characteristic features) and methods (things they can do). For example, an object car could be defined to have the properties model, make, color, number of doors, position etc., and the methods steer right, steer left, accelerate, break, stop, change gear, repaint,… whose application might affect the state of the object.
According to Python’s official documentation:
“Objects are Python’s abstraction for data. All data in a Python program is represented by objects or by relations between objects.”
All you need to know for now is that in Python objects have an identifier (ID) (i.e. their name), a type (numbers, text, collections,…) and a value (the actual data represented by the objects). Once an object has been created the identifier and the type never change, while its value can either change (mutable objects) or stay constant (immutable objects).
Python provides the following built-in data types:
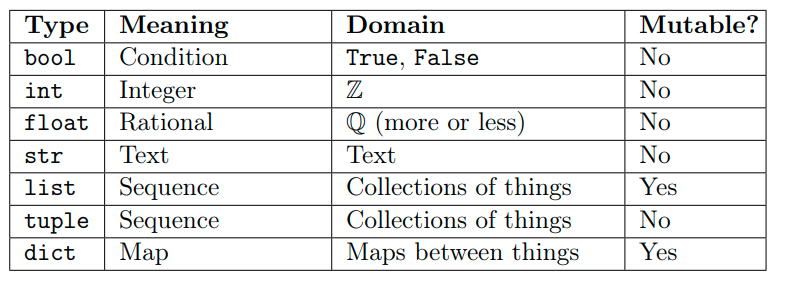
We will stick with the simplest ones for now, but later on we will dive deeper into all of them.
Variables¶
Variables are just references to objects, in other words they are the name given to an object. Variables can be assigned to objects by using the assignment operator =.
The instruction
[2]:
sides = 4
might represent the number of sides of a square. What happens when we execute it in Python? An object is created, it is given an identifier, its type is set to “int” (an integer number), its value to 4 and a name sides is placed in the current namespace to point to that object, so that after that instruction we can access that object through its name. The type of an object can be accessed with the function type() and the identifier with the function id():
[3]:
sides = 4
print( type(sides) )
print( id(sides) )
<class 'int'>
10914592
Consider now the following code:
[4]:
sides = 4 #a square
print ("value:", sides, " type:", type(sides), " id:", id(sides))
sides = 5 #a pentagon
print ("value:", sides, " type:", type(sides), " id:", id(sides))
value: 4 type: <class 'int'> id: 10914592
value: 5 type: <class 'int'> id: 10914624
The value of the variable sides has been changed from 4 to 5, but as stated in the table above, the type int is immutable. Luckily, this did not prevent us to change the value of sides from 4 to 5. What happened behind the scenes when we executed the instruction sides = 5 is that a new object has been created of type int (5 is still an integer) and it has been made accessible with the same name sides, but since it is a different object (i.e. the integer 5). As a poof of this,
check that the that the identifier printed above is actually different.
Note: You do not have to really worry about what happens behind the scenes, as the Python interpreter will take care of these aspects for you, but it is nice to know what it does.
You can even change the type of a variable during execution but that is normally a bad idea as it makes understanding the code more complicated and leaves more room for errors.
Python allows you to do (but, please, REFRAIN FROM DOING SO!):
[5]:
sides = 4 #a square
print ("value:", sides, " type:", type(sides), " id:", id(sides))
sides = "four" #the sides in text format
print ("value:", sides, " type:", type(sides), " id:", id(sides))
value: 4 type: <class 'int'> id: 10914592
value: four type: <class 'str'> id: 140640184741312
IMPORTANT NOTE: You can choose the name that you like for your variables (I advise to pick something reminding their meaning), but you need to adhere to some simple rules.
Names can only contain upper/lower case digits (
A-Z,a-z), numbers (0-9) or underscores_;Names cannot start with a number;
Names cannot be equal to reserved keywords:

Numeric types¶
We already mentioned that numbers are immutable objects. Python provides different numeric types: integers, booleans, reals (floats) and even complex numbers and fractions (but we will not get into those).
Integers¶
Their range of values is limited only by the memory available. As we have already seen, python provides also a set of standard operators to work with numbers:
[6]:
a = 7
b = 4
print(a + b) # 11
print(a - b) # 3
print(a // b) # integer division: 1
print(a * b) # 28
print(a ** b) # power: 2401
print(a / b) # division 1.75
print(type(a / b))
11
3
1
28
2401
1.75
<class 'float'>
Note that in the latter case the result is no more an integer, but a float (we will get to that later).
Booleans¶
These objects are used for the boolean algebra. Truth values are represented with the keywords True and False in Python. A boolean object can only have value True or False. We can convert booleans into integers with the builtin function int. Any integer can be converted into a boolean (and vice-versa) with:
[7]:
a = bool(1)
b = bool(0)
c = bool(72)
d = bool(-5)
t = int(True)
f = int(False)
print("a: ", a, " b: ", b, " c: ", c, " d: ", d , " t: ", t, " f: ", f)
a: True b: False c: True d: True t: 1 f: 0
any integer is evaluated to true, except 0. Note that, the truth values True and False respectively behave like the integers 1 and 0.
We can operate on boolean values with the boolean operators and, or, not. Recall boolean algebra for their use:
[8]:
T = True
F = False
print ("T: ", T, " F:", F)
print ("T and F: ", T and F) #False
print ("T and T: ", T and T) #True
print ("F and F: ", F and F) #False
print ("not T: ", not T) # False
print ("not F: ", not F) # True
print ("T or F: ", T or F) # True
print ("T or T: ", T or T) # True
print ("F or F: ", F or F) # False
T: True F: False
T and F: False
T and T: True
F and F: False
not T: False
not F: True
T or F: True
T or T: True
F or F: False
Numeric comparators are operators that return a boolean value. Here are some examples:
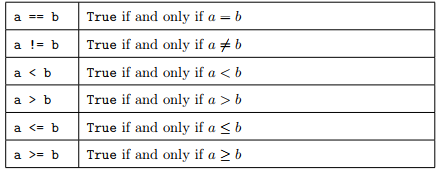
Example: Given a variable a = 10 and a variable b = 77, let’s swap their values (i.e. at the end a will be equal to 77 and b to 10). Let’s also check the values at the beginning and at the end.
[9]:
a = 10
b = 77
print("a: ", a, " b:", b)
print("is a equal to 10?", a == 10)
print("is b equal to 77?", b == 77)
TMP = b #we need to store the value of b safely
b = a #ok, the old value of b is gone... is it?
a = TMP #a gets the old value of b... :-)
print("a: ", a, " b:", b)
print("is a equal to 10?", a == 10)
print("is a equal to 77?", a == 77)
print("is b equal to 10?", b == 10)
print("is b equal to 77?", b == 77)
a: 10 b: 77
is a equal to 10? True
is b equal to 77? True
a: 77 b: 10
is a equal to 10? False
is a equal to 77? True
is b equal to 10? True
is b equal to 77? False
Real numbers¶
Python stores real numbers (floating point numbers) in 64 bits of information divided in sign, exponent and mantissa.
Example: Let’s calculate the area of the center circle of a football pitch (radius = 9.15m) recalling that \(area= \Pi*R^2\):
[10]:
R = 9.15
Pi = 3.141592653589793
Area = Pi*(R**2)
print (Area)
263.02199094017146
Note that the builtin math module of python contains the definition of \(\Pi\), therefore we could rewrite the code above as:
[11]:
import math
R = 9.15
Pi = math.pi
Area = Pi*(R**2)
print (Area)
263.02199094017146
Note that the parenthesis around the R**2 are not necessary as the operator ** has the precedence, but I personally think it helps readability.
Here is a reminder of the precedence of operators:
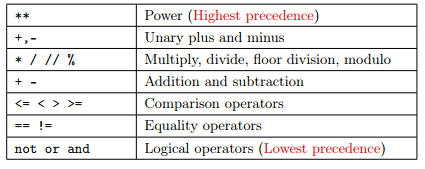
Example: Let’s compute the GC content of a DNA sequence 33 base pairs long, having 12 As, 9 Ts, 5 Cs and 7Gs. The GC content can be expressed by the formula: \(gc = \frac{G+C}{A+T+C+G}\) where A,T,C,G represent the number of nucleotides of each kind. What is the AT content? Is the GC content higher than the AT content?
[12]:
A = 12
T = 9
C = 5
G = 7
gc = (G+C)/(A+T+C+G)
print("The GC content is: ", gc)
at = 1 - gc
print("The AT content is: ", at)
print (gc > at)
The GC content is: 0.36363636363636365
The AT content is: 0.6363636363636364
False
Strings¶
Strings are immutable objects (note the actual type is str) used by python to handle text data. Strings are sequences of unicode code points that can represent characters, but also formatting information (e.g. ‘\n’ for new line). Unlike other programming languages, python does not have the data type character, which is represented as a string of length 1.
There are several ways to define a string:
[13]:
S = "my first string, in double quotes"
S1 = 'my second string, in single quotes'
S2 = '''my third string is
in triple quotes
therefore it can span several lines'''
S3 = """my fourth string, in triple double-quotes
can also span
several lines"""
print(S, '\n') #let's add a new line at the end of the string with \n
print(S1,'\n')
print(S2, '\n')
print(S3, '\n')
my first string, in double quotes
my second string, in single quotes
my third string is
in triple quotes
therefore it can span several lines
my fourth string, in triple double-quotes
can also span
several lines
To put special characters like ‘,” and so on you need to “escape them” (i.e. write them following a back-slash).

Example: Let’s print a string containing a quote and double quote (i.e. ‘ and “).
[14]:
myString = "This is how I \'quote\' and \"double quote\" things in strings"
print(myString)
This is how I 'quote' and "double quote" things in strings
Strings can be converted to and from numbers with the functions str(), int() or float().
Example: Let’s define a string myString with the value “47001” and convert it into an int. Try adding one and print the result.
[15]:
my_string = "47001"
print(my_string, " has type ", type(my_string))
my_int = int(my_string)
print(my_int, " has type ", type(my_int))
my_int = my_int + 1 #adds one
my_string = my_string + "1" #cannot add 1 (we need to use a string).
#This will append 1 at the end of the string
print(my_int)
print(my_string)
47001 has type <class 'str'>
47001 has type <class 'int'>
47002
470011
Be careful though that if the string cannot be converted into an integer, then you get an error
[16]:
my_wrong_number = "13a"
N = int(my_wrong_number)
print(N)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-16-bcfe98c1ea66> in <module>
1 my_wrong_number = "13a"
2
----> 3 N = int(my_wrong_number)
4
5 print(N)
ValueError: invalid literal for int() with base 10: '13a'
Python defines some operators to work with strings. Recall the slides shown during the lecture:
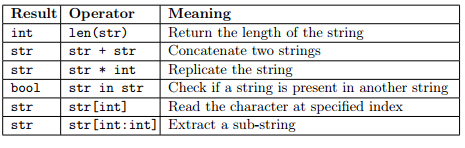
Example A tandem repeat is a short sequence of DNA that is repeated several times in a row. Let’s create a string representing the tandem repeat of the motif “ATTCG” repeated 5 times. What is the length of the whole repetitive region? Is the motif “TCGAT” (m1) present in the region? The motif “TCCT” (m2)? Let’s give an orientation to the tandem repeat by adding the string “5’-” (5’ end) on the left and “-3’” (3’ end) to the right.
[17]:
motif = "ATTCG"
tandem_repeat = motif * 5
print(motif)
print(tandem_repeat, " has length", len(tandem_repeat))
m1 = "TCGAT"
m2 = "TCCT"
print("Is ", m1, " in ", tandem_repeat, " ? ", m1 in tandem_repeat )
print("Is ", m2, " in ", tandem_repeat, " ? ", m2 in tandem_repeat )
oriented_tr = "5\'-" + tandem_repeat + "-3\'"
print(oriented_tr)
ATTCG
ATTCGATTCGATTCGATTCGATTCG has length 25
Is TCGAT in ATTCGATTCGATTCGATTCGATTCG ? True
Is TCCT in ATTCGATTCGATTCGATTCGATTCG ? False
5'-ATTCGATTCGATTCGATTCGATTCG-3'
We can access strings at specific positions (indexing) or get a substring starting from a position S to a position E. The only thing to remember is that numbering starts from 0. Thei-th character of a string can be accessed as str[i-1]. Substrings can be accessed as str[S:E], optionally a third parameter can be specified to set the step (i.e. str[S:E:STEP]).
Important note. Remember that when you do str[S:E], S is inclusive, while E is exclusive (see S[0:6] below).
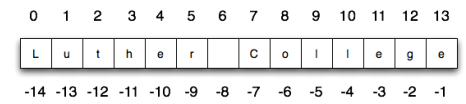
Let’s see these aspects in action with an example:
[18]:
S = "Luther College"
print(S) #print the whole string
print(S == S[:]) #a fancy way of making a copy of the original string
print(S[0]) #first character
print(S[3]) #fourth character
print(S[-1]) #last character
print(S[0:6]) #first six characters
print(S[-7:]) #final seven characters
print(S[0:len(S):2]) #every other character starting from the first
print(S[1:len(S):2]) #every other character starting from the second
Luther College
True
L
h
e
Luther
College
Lte olg
uhrClee
Methods for the str object¶
The object str has some methods that can be applied to it (remember methods are things you can do on objects). Recall from the lecture that the main methods are:
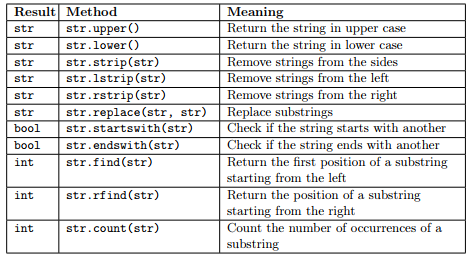
IMPORTANT NOTE: Since Strings are immutable, every operation that changes the string actually produces a new str object having the modified string as value.
Moreover, since strings are immutable we cannot directly change them with an assignment operator.
Example: Since the genetic code is degenerate, there are many codons encoding for the same aminoacid. Consider Proline, it can be encoded by the following codons: CCU, CCA,CCG, CCC. Let’s create a string proline and assign it to its possible codons one after the other.
[19]:
"""
Wrong solution. We cannot directly replace the value of a string
"""
proline = "CCU"
print("Proline can be encoded by: ", proline)
proline[2]="A"
print(".. or by: ", proline)
Proline can be encoded by: CCU
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-19-9750dcfa1cbd> in <module>
5 proline = "CCU"
6 print("Proline can be encoded by: ", proline)
----> 7 proline[2]="A"
8 print(".. or by: ", proline)
9
TypeError: 'str' object does not support item assignment
[20]:
"""
Correct solution. Using str.replace
"""
proline = "CCU"
print("Proline can be encoded by: ", proline)
proline = proline.replace("U","A")
print(".. or by: ", proline)
proline = proline.replace("A","G")
print(".. or by: ", proline)
proline = proline.replace("G","C")
print(".. or by: ", proline)
Proline can be encoded by: CCU
.. or by: CCA
.. or by: CCG
.. or by: CCC
[21]:
"""
Another correct solution. Using string slicing and catenation.
"""
proline = "CCU"
print("Proline can be encoded by: ", proline)
proline = proline[:-1]+"A" #equal to proline[0:-1] or proline[0:2]
print(".. or by: ", proline)
proline = proline[:-1]+"G"
print(".. or by: ", proline)
proline = proline[:-1]+"C"
print(".. or by: ", proline)
Proline can be encoded by: CCU
.. or by: CCA
.. or by: CCG
.. or by: CCC
Example: Given the DNA sequence S = ” aTATGCCCATatcgctAAATTGCTGCCATTACA “. Print its length (removing any blank spaces at either sides), the number of adenines, cytosines, guanines and thymines present. Is the sequence “ATCG” present in S? Print how many times the substring “TGCC” appears in S and all the corresponding indexes.
[22]:
S = " aTATGCCCATatcgctAAATTGCTGCCATTACA "
print(S)
S = S.strip(" ")
print(S)
print(len(S))
tmp_s = S.upper() #for simplicity to count only 4 different nucleotides
print("A count: ", tmp_s.count("A"))
print("C count: ", tmp_s.count("C"))
print("T count: ", tmp_s.count("T"))
print("G count: ", tmp_s.count("G"))
print("Is ATCG in ", tmp_s, "? ", tmp_s.find("ATCG") != -1) #or tmp_s.count("ATCG") > 0
print("TGCC is present ", tmp_s.count("TGCC"), " times in ", tmp_s)
print("TGCC is present at pos ", tmp_s.find("TGCC"))
print("TGCC is present at pos ", tmp_s.rfind("TGCC")) #or tmp_S.find("TGCC",4)
aTATGCCCATatcgctAAATTGCTGCCATTACA
aTATGCCCATatcgctAAATTGCTGCCATTACA
33
A count: 10
C count: 9
T count: 10
G count: 4
Is ATCG in ATATGCCCATATCGCTAAATTGCTGCCATTACA ? True
TGCC is present 2 times in ATATGCCCATATCGCTAAATTGCTGCCATTACA
TGCC is present at pos 3
TGCC is present at pos 23
Exercises¶
Given the following string on two lines:
text = """Nobody said it was easy
No one ever said it would be this hard"""
write some python code that a)prints the whole string; b) prints the first and last character; c) prints the first 10 characters; d) prints from the 19th character to the 31st; e) prints the string all in capital letters.
Show/Hide Solution
An exon of a gene starts from position 12030 on a genome and ends at position 12174. Does an A/T SNP present at position 12111 affect this exon (i.e. is it inside the exon)? And what about a SNP present at position 12188? Hint: create a suitable boolean expression to check if the positions are within the interval of the exon.
Show/Hide Solution
SNP FB_AFFY_0000024 of the Apple 480K SNP chip has 5’ flanking region (i.e. the forward probe) CATTATTTTCACTTGGGTCGAGGCCAGATTCCATC and 3’ flanking region (i.e. the reverse probe) GGATTGCCCGAAATCAGAGAAAAGTCG. The SNP is a G/A transversion. Answer the following questions:
What is the length of the 5’ flanking region? And that of the 3’ flanking region?
The IUPAC code of the G/A transversion is R. What is the sequence of the whole region using the “[G/A]” notation for the SNP (hint: concatenate it in a new string called region) and the iupac notation R (region_iupac)?
Retrive and print only the SNP from region and iupac_region
Show/Hide Solution
Compute the melting temperature \(T_m\) of the primer with sequence “TTAGCACACGTGAGCCAATGGAGCAAACGGGTAATT”. The melting temperature \(T_m\) (in degrees Celtius) can be computed as: \(T_m = 64.9 + 41(GC - 16.4)/N\), where \(GC\) is the total number of G and C in the primer and \(N\) is its length.
Show/Hide Solution
The spike protein of the Sars-CoV-2 virus has the following aminoacidic sequence:
S = """ MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFS NVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIV NNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLE GKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQT LLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETK CTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISN CVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIAD YNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPC NGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVN FNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITP GTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSY ECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTI SVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQE VFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDC LGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAM QMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALN TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPA ICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDP LQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDL QELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDD SEPVLKGVKLHYT """
Write a little python script to answer the following questions: 1) What are the first 10 and the last 10 aminoacids? 2) How many aminoacids does it have (beware of new lines)? 3) How many Tyrosines (T) does it contain? 4) How many Triptophanes (W)? 5) How many Valines (V) followed by at least one Lysine (K)?
Show/Hide Solution
Convert the following extract of the PalB2 gene into mRNA (i.e. replace thymine with uracile):
seq ="""CTGTCTCCCTCACTGTATGTAAATTGCATCTAGAATAGCA
TCTGGAGCACTAATTGACACATAGTGGGTATCAATTATTA
TTCCAGGTACTAGAGATACCTGGACCATTAACGGATAAAT
AGAAGATTCATTTGTTGAGTGACTGAGGATGGCAGTTCCT
GCTACCTTCAAGGATCTGGATGATGGGGAGAAACAGAGAA
CATAGTGTGAGAATACTGTGGTAAGGAAAGTACAGAGGAC
TGGTAGAGTGTCTAACCTAGATTTGGAGAAGGACCTAGAA
GTCTATCCCAGGGAAATAAAAATCTAAGCTAAGGTTTGAG
GAATCAGTAGGAATTGGCAAAGGAAGGACATGTTCCAGAT
GATAGGAACAGGTTATGCAAAGATCCTGAAATGGTCAGAG
CTTGGTGCTTTTTGAGAACCAAAAGTAGATTGTTATGGAC
CAGTGCTACTCCCTGCCTCTTGCCAAGGGACCCCGCCAAG
CACTGCATCCCTTCCCTCTGACTCCACCTTTCCACTTGCC
CAGTATTGTTGGTGT"""
and print the number of uracils present and the total length of the sequence (remember to remove newlines).
Considering the genetic code and all the possible open reading frames, answer the following questions:
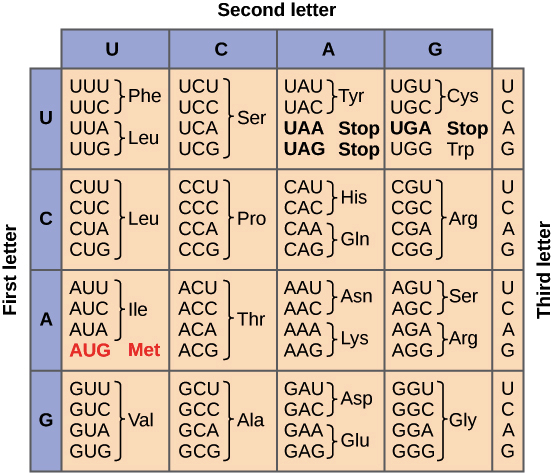
How many stop codons are present in the sequence?
How many Glycines (Gly)?
Is Tryptophane (Trp) present?
What is the position of the leftmost Trp? Print the codon to double check correctness (hint: slicing).
What is the position of the rightmost Trp? Print the codon to double check correctness (hint: slicing).
Show/Hide Solution
Consider the following Illumina HiSeq 4000 read:
read = """AATGATACGGCGACCACCGAGATCTACACGCCTCCCTCGCGC
CATCAGAGAGTCTGGGTCTCAGGTACCGCAGTTGTATCTTGCGCGACTATA
ATCCACGGCTCTTATTCTAGCGTGCGCGTACGGCGGTGGGCGTCGTTACGCTATATT"""
and try to answer the following questions:
1. How long is the read (beware of newlines)?
2. What is the GC content of the read (remember $gc = \frac{G+C}{A+T+C+G}$)?
3. A Nextera adapter is "AATGATACGGCGACCACCGAGATCTACACGCCTCCCTCGCGCCATCAG".
Is it present in the read? How long is it?
4. Remove the Nextera adapter from the read and recompute the GC content.
Has GC content increased after adapter trimmming?
Show/Hide Solution
Given geneA starting at position 1000 and ending at position 3400, and geneB starting at position 3700 and ending at position 6000. Randomly select a position (pos) from 1 to 5202 and check the following: a. is pos in geneA? b. is pos in geneB? c. is pos in between the two genes? d. is pos within one of the two genes? e. is pos outside both genes? f. is pos within 100 bases before the start of geneA? To pick a random number you can import the random module and use the random.randint(start,end) function:
import randompos = random.randint(1,6000)
Show/Hide Solution
The DNA-binding domain of the Tumor Suppressor Protein TP53 can be represented by the string:
chain_a = """SSSVPSQKTYQGSYGFRLGFLHSGTAKSVTCTYSPALNKM
FCQLAKTCPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVV
RRCPHHERCSDSDGLAPPQHLIRVEGNLRVEYLDDRNTFR
HSVVVPYEPPEVGSDCTTIHYNYMCNSSCMGGMNRRPILT
IITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKG
EPHHELPPGSTKRALPNNT"""
Answer the following questions:
1. How many lines is the sequence written on?
2. How long is the sequence (remove newlines)?
3. Create a new sequence with all new lines removed
4. How many cysteines "C" and histidines "H" are there in the sequence?
5. Does the chain contain the sub-sequence "NLRVEYLDDRN"? Where?
6. Extract the first line of the sequence (Hint: use find and string slicing).
Show/Hide Solution
Calculate the zeros of the equation \(ax^2-b = 0\) where a = 10 and b = 1. Hint: use math.sqrt or ** 0.5. Finally check that substituting the obtained value of x in the equation gives zero.
Show/Hide Solution