Practical 5¶
In this practical we will add some information on loops and introduce dictionaries.
More on loops¶
As seen in the previous practical and in the lecture, there are three different ways of execution flow:
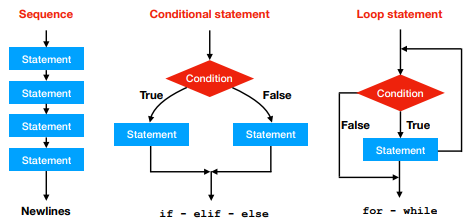
We have already seen the if, for and while loops and their variants. Please remember that the code block of each of these statements is defined by the indentation (i.e. spacing).
Ternary operator¶
In some cases it is handy to be able to initialize a variable depending on the value of another one.
Example: The discount rate applied to a purchase depends on the amount of the sale. Create a variable discount setting its value to 0 if the variable amount is lower than 100 euros, to 10% if it is higher.
[1]:
amount = 110
discount = 0
if amount >100:
discount = 0.1
else:
discount = 0 # not necessary
print("Total amount:", amount, "discount:", discount)
Total amount: 110 discount: 0.1
The previous code can be written more coincisely as:
[2]:
amount = 110
discount = 0.1 if amount > 100 else 0
print("Total amount:", amount, "discount:", discount)
Total amount: 110 discount: 0.1
The basic syntax of the ternary operator is:
variable = value if condition else other_value
meaning that the variable is initialized to value if the condition holds, otherwise to other_value.
Python also allows in line operations separated by a “;”. This is a compact way of specifying several instructions on a single line, but it makes the code more difficult to read.
[3]:
a = 10; b = a + 1; c = b +2
print(a,b,c)
10 11 13
Note: Although the ternary operator and in line operations are sometimes useful and less verbose than the explicit definition, they are considered “non-pythonic” and advised against.
Break and continue¶
Sometimes it is useful to skip an entire iteration of a loop or end the loop before its supposed end. This can be achieved with two different statements: continue and break.
Continue statement¶
Within a for or while loop, continue makes the interpreter skip that iteration and move on to the next.
Example: Print all the odd numbers from 1 to 20.
[4]:
#Two equivalent ways
#1. Testing remainder == 1
for i in range(21):
if i % 2 == 1:
print(i, end = " ")
print("")
#2. Skipping if remainder == 0 in for
for i in range(21):
if i % 2 == 0:
continue
print(i, end = " ")
1 3 5 7 9 11 13 15 17 19
1 3 5 7 9 11 13 15 17 19
Continue can be used also within while loops but we need to be careful to update the value of the variable before reaching the continue statement or we will get stuck in never-ending loops. Example: Print all the odd numbers from 1 to 20.
[ ]:
#Wrong code:
i = 0
while i < 21:
if i % 2 == 0:
continue
print(i, end = " ")
i = i + 1 # NEVER EXECUTED IF i % 2 == 0!!!!
a possible correct solution using while:
[5]:
i = -1
while i< 20: #i is incremented in the loop, so 20!!!
i = i + 1 #the variable is updated no matter what
if i % 2 == 0:
continue
print(i, end = " ")
1 3 5 7 9 11 13 15 17 19
Break statement¶
Within a for or while loop, break makes the interpreter exit the loop and continue with the sequential execution. Sometimes it is useful to get out of the loop if to complete our task we do not need to get to the end of it.
Example: Given the following list of integers [1,5,6,4,7,1,2,3,7] print them until a number already printed is found.
[6]:
L = [1,5,6,4,7,1,2,3,7]
found = []
for i in L:
if i in found:
break
found.append(i)
print(i, end = " ")
1 5 6 4 7
Example: Pick a random number from 1 and 50 and count how many times it takes to randomly choose number 27. Limit the number of random picks to 40 (i.e. if more than 40 picks have been done and 27 has not been found exit anyway with a message).
[7]:
import random
iterations = 1
picks = []
while iterations <= 40:
pick = random.randint(1,50)
picks.append(pick)
if pick == 27:
break
iterations += 1 #equal to: iterations = iterations + 1
if iterations == 41:
print("Sorry number 27 was never found!")
else:
print("27 found in ", iterations, "iterations")
print(picks)
27 found in 9 iterations
[9, 16, 8, 37, 42, 7, 14, 28, 27]
An alternative way without using the break statement makes use of a flag variable (that when changes value will make the loop end):
[8]:
import random
found = False # This is called flag
iterations = 1
picks = []
while iterations <= 40 and found == False: #the flag is used to exit
pick = random.randint(1,50)
picks.append(pick)
if pick == 27:
found = True #update the flag, will exit at next iteration
iterations += 1
if iterations == 41 and not found:
print("Sorry number 27 was never found!")
else:
print("27 found in ", iterations -1, "iterations")
print(picks)
Sorry number 27 was never found!
[14, 32, 28, 25, 49, 28, 31, 6, 11, 17, 21, 13, 35, 15, 3, 30, 34, 19, 38, 20, 47, 38, 14, 42, 32, 19, 23, 49, 40, 21, 17, 35, 47, 1, 39, 41, 31, 33, 21, 35]
List comprehension¶
List comprehension is a quick way of creating a list. The resulting list is normally obtained by applying a function or a method to the elements of another list that remains unchanged.
The basic syntax is:
new_list = [ some_function (x) for x in start_list]
or
new_list = [ x.some_method() for x in start_list]
List comprehension can also be used to filter elements of a list and produce another list as sublist of the first one (remember that the original list is not changed).
In this case the syntax is:
new_list = [ some_function (x) for x in start_list if condition]
or
new_list = [ x.some_method() for x in start_list if condition]
where the element x in start_list becomes part of new_list if and only if the condition holds True.
Let’s see some examples:
Example: Given a list of strings [“hi”, “there”, “from”, “python”] create a list with the length of the corresponding element (i.e. the one with the same index).
[9]:
elems = ["hi", "there", "from", "python"]
newList = [len(x) for x in elems]
for i in range(0,len(elems)):
print(elems[i], " has length ", newList[i])
hi has length 2
there has length 5
from has length 4
python has length 6
Example: Given a list of strings [“dog”, “cat”, “rabbit”, “guinea pig”, “hamster”, “canary”, “goldfish”] create a list with the elements starting with a “c” or “g”.
[10]:
pets = ["dog", "cat", "rabbit", "guinea pig", "hamster", "canary", "goldfish"]
cg_pets = [x for x in pets if x.startswith("c") or x.startswith("g")]
print("Original:")
print(pets)
print("Filtered:")
print(cg_pets)
Original:
['dog', 'cat', 'rabbit', 'guinea pig', 'hamster', 'canary', 'goldfish']
Filtered:
['cat', 'guinea pig', 'canary', 'goldfish']
Example: Create a list with all the numbers divisible by 17 from 1 to 200.
[11]:
values = [ x for x in range(1,200) if x % 17 == 0]
print(values)
[17, 34, 51, 68, 85, 102, 119, 136, 153, 170, 187]
Example: Transpose the matrix \(\begin{bmatrix}1 & 10\\2 & 20\\3 & 30\\4 & 40\end{bmatrix}\) stored as a list of lists (i.e. matrix = [[1, 10], [2,20], [3,30], [4,40]]). The output matrix should be: \(\begin{bmatrix}1 & 2 & 3 & 4\\10 & 20 & 30 & 40\end{bmatrix}\), represented as [[1, 2, 3, 4], [10, 20, 30, 40]]
[12]:
matrix = [[1, 10], [2,20], [3,30], [4,40]]
print(matrix)
transpose = [[row[i] for row in matrix] for i in range(2)]
print (transpose)
[[1, 10], [2, 20], [3, 30], [4, 40]]
[[1, 2, 3, 4], [10, 20, 30, 40]]
Example: Given the list: [“Hotel”, “Icon”,” Bus”,”Train”, “Hotel”, “Eye”, “Rain”, “Elephant”] create a list with all the first letters.
[13]:
myList = ["Hotel", "Icon"," Bus","Train", "Hotel", "Eye", "Rain", "Elephant"]
initials = [x[0] for x in myList]
print(myList)
print(initials)
print("".join(initials))
['Hotel', 'Icon', ' Bus', 'Train', 'Hotel', 'Eye', 'Rain', 'Elephant']
['H', 'I', ' ', 'T', 'H', 'E', 'R', 'E']
HI THERE
With list comprehension we can copy a list into another one, but this is a shallow copy:
[10]:
a = [1,2,3]
b = [a, [[a]]]
print("B:", b)
c = [x for x in b]
print("C:", c)
a.append(4)
print("B now:" , b)
print("C now:", c)
B: [[1, 2, 3], [[[1, 2, 3]]]]
C: [[1, 2, 3], [[[1, 2, 3]]]]
B now: [[1, 2, 3, 4], [[[1, 2, 3, 4]]]]
C now: [[1, 2, 3, 4], [[[1, 2, 3, 4]]]]
Dictionaries¶
A dictionary is a map between one object, the key, and another object, the value. Dictionaries are mutable objects and contain sequences of mappings key –> object but there is not specific ordering among them. Dictionaries are defined using the curly braces {key1 : value1, key2 : value2} and : to separate keys from values.
Some examples on how to define dictionaries follow:
[14]:
first_dict = {"one" : 1, "two": 2, "three" : 3, "four" : 4}
print("First:", first_dict)
empty_dict = dict()
print("Empty:",empty_dict)
second_dict = {1 : "one", 2 : "two", "three" :3 } #BAD IDEA BUT POSSIBLE!!!
print(second_dict)
third_dict = dict(zip(["one","two","three","four"],[1,2,3,4]))
print(third_dict)
print(first_dict == third_dict)
First: {'one': 1, 'two': 2, 'three': 3, 'four': 4}
Empty: {}
{1: 'one', 2: 'two', 'three': 3}
{'one': 1, 'two': 2, 'three': 3, 'four': 4}
True
Note that there is no ordering of the keys, and that the order in which they have been inserted is not preserved. Moreover, keys and values can be dishomogeneous (e.g. keys can be strings and values integers). An interesting case is third_dict where the function zip followed by dict is used to map the keys of the first list into the values present in the second.
Note that keys can be dishomogeneous, even though this is a bad idea normally. The only requirement for the keys is that they must be immutable objects. Trying to use a mutable object as a key will make the interpreter crash with the error: unhashable type. Finally, keys must be unique. We cannot associate more than one value to the same key.
[15]:
a = (1,2,3) #a,b are tuples: hence immutable
b = (1,3,5)
my_dict = {a : 6, b : 9 }
print(my_dict)
c = [1,2,3] #c,d are lists: hence mutable
d = [1,3,5]
dict2 = {c : 6, d : 9}
print(dict2)
{(1, 2, 3): 6, (1, 3, 5): 9}
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-15-0fe98c7f5acd> in <module>
8 d = [1,3,5]
9
---> 10 dict2 = {c : 6, d : 9}
11 print(dict2)
TypeError: unhashable type: 'list'
Functions working on dictionaries¶
As for the other data types, python provides several operators that can be applied to dictionaries. The following operators are available and they basically work as in lists. The only exception being that the operator in checks whether the specified object is present among the keys.
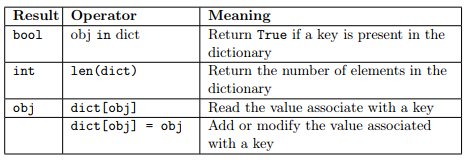
Some usage examples follow:
[16]:
myDict = {"one" : 1, "two" : 2, "twentyfive" : 25}
print(myDict)
myDict["ten"] = 10
myDict["twenty"] = 20
print(myDict)
myDict["ten"] = "10-again"
print(myDict)
print("The dictionary has ", len(myDict), " elements")
print("The value of \"ten\" is:", myDict["ten"])
print("The value of \"two\" is:", myDict["two"])
print("Is \"twentyfive\" in dictionary?", "twentyfive" in myDict)
print("Is \"seven\" in dictionary?", "seven" in myDict)
{'one': 1, 'two': 2, 'twentyfive': 25}
{'one': 1, 'two': 2, 'twentyfive': 25, 'ten': 10, 'twenty': 20}
{'one': 1, 'two': 2, 'twentyfive': 25, 'ten': '10-again', 'twenty': 20}
The dictionary has 5 elements
The value of "ten" is: 10-again
The value of "two" is: 2
Is "twentyfive" in dictionary? True
Is "seven" in dictionary? False
Dictionary methods¶
Recall what seen in the lecture, the following methods are available for dictionaries:
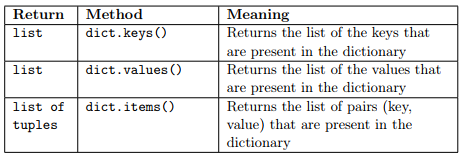
These methods are new to dictionaries and can be used to loop through the elements in them.
ERRATUM: dict.keys() returns a dict_keys object not a list. To cast it to list, we need to call list(dict.keys()). The same applies to dict.values() that returns a dict_values object that needs conversion to a list with list(dict.values()).
[17]:
D = {"k1" : 1, "k2" : 2 , "k3" : 3}
print("keys:" , D.keys(), "values:", D.values())
print("")
print("keys:", list(D.keys()), "values:", list(D.values()))
keys: dict_keys(['k1', 'k2', 'k3']) values: dict_values([1, 2, 3])
keys: ['k1', 'k2', 'k3'] values: [1, 2, 3]
Example Given the protein sequence below, store in a dictionary all the aminoacids present and count how many times they appear. Finally print out the stats (e.g. how many amino-acids are present, the most frequent, the least frequent and the frequency of all of them in alphabetical order).
>sp|P00517|KAPCA_BOVIN cAMP-dependent protein kinase catalytic subunit alpha
MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVML
VKHMETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMV
MEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY
IQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF
ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT
TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF
[1]:
protein = """MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVML
VKHMETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMV
MEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY
IQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF
ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT
TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF"""
protein = protein.replace("\n","")
print(protein)
amino_acids = dict()
for a in protein:
if a in amino_acids:
amino_acids[a] = amino_acids[a] + 1 # amino_acids[a] += 1
else:
amino_acids[a] = 1
num_aminos = len(amino_acids)
print("The number of amino-acids present is ", num_aminos)
#let's get all aminoacids
#and sort them alphabetically
a_keys = list(amino_acids.keys())
a_keys.sort()
# Another example of dictionaries
mostF = {"frequency" : -1, "aminoacid" : "-"}
leastF = {"frequency" : len(protein), "aminoacid" : "-"}
for a in a_keys:
freq = amino_acids[a]
if(mostF["frequency"] < freq):
mostF["frequency"] = freq
mostF["aminoacid"] = a
if(leastF["frequency"] > freq):
leastF["frequency"] = freq
leastF["aminoacid"] = a
print(a, " is present", freq, "times")
print("Amino", leastF["aminoacid"], "has the lowest freq. (",leastF["frequency"],")")
print("Amino", mostF["aminoacid"], "has the highest freq. (",mostF["frequency"],")")
MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHMETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF
The number of amino-acids present is 20
A is present 23 times
C is present 2 times
D is present 18 times
E is present 27 times
F is present 25 times
G is present 22 times
H is present 9 times
I is present 21 times
K is present 34 times
L is present 32 times
M is present 8 times
N is present 17 times
P is present 14 times
Q is present 14 times
R is present 15 times
S is present 16 times
T is present 14 times
V is present 20 times
W is present 6 times
Y is present 14 times
Amino C has the lowest freq. ( 2 )
Amino K has the highest freq. ( 34 )
Important NOTE. Accessing a value through the key of a dictionary requires that the pair key-value one searches for is present in the dictionary. If the searched key is not present the interpreter crashes out throwing a KeyError as follows:
[19]:
myDict = {"one" : 1, "two" : 2, "three" : 3}
print(myDict["one"])
print(myDict["seven"])
1
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
<ipython-input-19-a05b31e54a02> in <module>
2
3 print(myDict["one"])
----> 4 print(myDict["seven"])
KeyError: 'seven'
This could be avoided by checking that the key is present in the dictionary beforehand:
[20]:
myDict = {"one" : 1, "two" : 2, "three" : 3}
search_keys = ["one", "seven"]
for s in search_keys:
if s in myDict:
print("key:", s, "value:", myDict[s])
else:
print("key", s, "not found in dictionary")
key: one value: 1
key seven not found in dictionary
or by using the dictionary method get that has two inputs: the key and a default value to return in case key is not present in the dictionary:
[21]:
myDict = {"one" : 1, "two" : 2, "three" : 3}
search_keys = ["one", "seven"]
for s in search_keys:
print("key:", s, "value:", myDict.get(s, "not found"))
key: one value: 1
key: seven value: not found
Exercises¶
Given the following two lists of integers: [1, 13, 22, 7, 43, 81, 77, 12, 15,21, 84,100] and [44,32,7, 100, 81, 13, 1, 21, 71]:
Sort the two lists
Create a third list as intersection of the two lists (i.e. an element is in the intersection if it is present in both lists).
Print the three lists.
Show/Hide Solution
The sequence below is the Sars-Cov2 ORF1a polyprotein. 1. Count and print how many aminoacids it is composed of and 2. put in a dictionary all the indexes of the occurrences of the following four aminoacids:
TTTL, GFAV, KMLL(i.e. the key of the dictionary is the sequence and the value is the list of all positions at which the four-mers appear).
ORF1a = """MESLVPGFNEKTHVQLSLPVLQVRDVLVRGFGDSVEEVLSEARQHLKDGTCGLVEVEKGVLPQLEQPYVF
IKRSDARTAPHGHVMVELVAELEGIQYGRSGETLGVLVPHVGEIPVAYRKVLLRKNGNKGAGGHSYGADL
KSFDLGDELGTDPYEDFQENWNTKHSSGVTRELMRELNGGAYTRYVDNNFCGPDGYPLECIKDLLARAGK
ASCTLSEQLDFIDTKRGVYCCREHEHEIAWYTERSEKSYELQTPFEIKLAKKFDTFNGECPNFVFPLNSI
IKTIQPRVEKKKLDGFMGRIRSVYPVASPNECNQMCLSTLMKCDHCGETSWQTGDFVKATCEFCGTENLT
KEGATTCGYLPQNAVVKIYCPACHNSEVGPEHSLAEYHNESGLKTILRKGGRTIAFGGCVFSYVGCHNKC
AYWVPRASANIGCNHTGVVGEGSEGLNDNLLEILQKEKVNINIVGDFKLNEEIAIILASFSASTSAFVET
VKGLDYKAFKQIVESCGNFKVTKGKAKKGAWNIGEQKSILSPLYAFASEAARVVRSIFSRTLETAQNSVR
VLQKAAITILDGISQYSLRLIDAMMFTSDLATNNLVVMAYITGGVVQLTSQWLTNIFGTVYEKLKPVLDW
LEEKFKEGVEFLRDGWEIVKFISTCACEIVGGQIVTCAKEIKESVQTFFKLVNKFLALCADSIIIGGAKL
KALNLGETFVTHSKGLYRKCVKSREETGLLMPLKAPKEIIFLEGETLPTEVLTEEVVLKTGDLQPLEQPT
SEAVEAPLVGTPVCINGLMLLEIKDTEKYCALAPNMMVTNNTFTLKGGAPTKVTFGDDTVIEVQGYKSVN
ITFELDERIDKVLNEKCSAYTVELGTEVNEFACVVADAVIKTLQPVSELLTPLGIDLDEWSMATYYLFDE
SGEFKLASHMYCSFYPPDEDEEEGDCEEEEFEPSTQYEYGTEDDYQGKPLEFGATSAALQPEEEQEEDWL
DDDSQQTVGQQDGSEDNQTTTIQTIVEVQPQLEMELTPVVQTIEVNSFSGYLKLTDNVYIKNADIVEEAK
KVKPTVVVNAANVYLKHGGGVAGALNKATNNAMQVESDDYIATNGPLKVGGSCVLSGHNLAKHCLHVVGP
NVNKGEDIQLLKSAYENFNQHEVLLAPLLSAGIFGADPIHSLRVCVDTVRTNVYLAVFDKNLYDKLVSSF
LEMKSEKQVEQKIAEIPKEEVKPFITESKPSVEQRKQDDKKIKACVEEVTTTLEETKFLTENLLLYIDIN
GNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKALRKVPTDNYITTYP
GQGLNGYTVEEAKTVLKKCKSAFYILPSIISNEKQEILGTVSWNLREMLAHAEETRKLMPVCVETKAIVS
TIQRKYKGIKIQEGVVDYGARFYFYTSKTTVASLINTLNDLNETLVTMPLGYVTHGLNLEEAARYMRSLK
VPATVSVSSPDAVTAYNGYLTSSSKTPEEHFIETISLAGSYKDWSYSGQSTQLGIEFLKRGDKSVYYTSN
PTTFHLDGEVITFDNLKTLLSLREVRTIKVFTTVDNINLHTQVVDMSMTYGQQFGPTYLDGADVTKIKPH
NSHEGKTFYVLPNDDTLRVEAFEYYHTTDPSFLGRYMSALNHTKKWKYPQVNGLTSIKWADNNCYLATAL
LTLQQIELKFNPPALQDAYYRARAGEAANFCALILAYCNKTVGELGDVRETMSYLFQHANLDSCKRVLNV
VCKTCGQQQTTLKGVEAVMYMGTLSYEQFKKGVQIPCTCGKQATKYLVQQESPFVMMSAPPAQYELKHGT
FTCASEYTGNYQCGHYKHITSKETLYCIDGALLTKSSEYKGPITDVFYKENSYTTTIKPVTYKLDGVVCT
EIDPKLDNYYKKDNSYFTEQPIDLVPNQPYPNASFDNFKFVCDNIKFADDLNQLTGYKKPASRELKVTFF
PDLNGDVVAIDYKHYTPSFKKGAKLLHKPIVWHVNNATNKATYKPNTWCIRCLWSTKPVETSNSFDVLKS
EDAQGMDNLACEDLKPVSEEVVENPTIQKDVLECNVKTTEVVGDIILKPANNSLKIIEEVGHTDLMAAYV
DNSSLTIKKPNELSRVLGLKTLATHGLAAVNSVPWDTIANYAKPFLNKVVSTTTNIVTRCLNRVCTNYMP
YFFTLLLQLCTFTRSTNSRIKASMPTTIAKNTVKSVGKFCLEASFNYLKSPNFSKLINIIIWFLLLSVCL
GSLIYSTAALGVLMSNLGMPSYCTGYREGYLNSTNVTIATYCTGSIPCSVCLSGLDSLDTYPSLETIQIT
ISSFKWDLTAFGLVAEWFLAYILFTRFFYVLGLAAIMQLFFSYFAVHFISNSWLMWLIINLVQMAPISAM
VRMYIFFASFYYVWKSYVHVVDGCNSSTCMMCYKRNRATRVECTTIVNGVRRSFYVYANGGKGFCKLHNW
NCVNCDTFCAGSTFISDEVARDLSLQFKRPINPTDQSSYIVDSVTVKNGSIHLYFDKAGQKTYERHSLSH
FVNLDNLRANNTKGSLPINVIVFDGKSKCEESSAKSASVYYSQLMCQPILLLDQALVSDVGDSAEVAVKM
FDAYVNTFSSTFNVPMEKLKTLVATAEAELAKNVSLDNVLSTFISAARQGFVDSDVETKDVVECLKLSHQ
SDIEVTGDSCNNYMLTYNKVENMTPRDLGACIDCSARHINAQVAKSHNIALIWNVKDFMSLSEQLRKQIR
SAAKKNNLPFKLTCATTRQVVNVVTTKIALKGGKIVNNWLKQLIKVTLVFLFVAAIFYLITPVHVMSKHT
DFSSEIIGYKAIDGGVTRDIASTDTCFANKHADFDTWFSQRGGSYTNDKACPLIAAVITREVGFVVPGLP
GTILRTTNGDFLHFLPRVFSAVGNICYTPSKLIEYTDFATSACVLAAECTIFKDASGKPVPYCYDTNVLE
GSVAYESLRPDTRYVLMDGSIIQFPNTYLEGSVRVVTTFDSEYCRHGTCERSEAGVCVSTSGRWVLNNDY
YRSLPGVFCGVDAVNLLTNMFTPLIQPIGALDISASIVAGGIVAIVVTCLAYYFMRFRRAFGEYSHVVAF
NTLLFLMSFTVLCLTPVYSFLPGVYSVIYLYLTFYLTNDVSFLAHIQWMVMFTPLVPFWITIAYIICIST
KHFYWFFSNYLKRRVVFNGVSFSTFEEAALCTFLLNKEMYLKLRSDVLLPLTQYNRYLALYNKYKYFSGA
MDTTSYREAACCHLAKALNDFSNSGSDVLYQPPQTSITSAVLQSGFRKMAFPSGKVEGCMVQVTCGTTTL
NGLWLDDVVYCPRHVICTSEDMLNPNYEDLLIRKSNHNFLVQAGNVQLRVIGHSMQNCVLKLKVDTANPK
TPKYKFVRIQPGQTFSVLACYNGSPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCVSFCYMHHMEL
PTGVHAGTDLEGNFYGPFVDRQTAQAAGTDTTITVNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKY
NYEPLTQDHVDILGPLSAQTGIAVLDMCASLKELLQNGMNGRTILGSALLEDEFTPFDVVRQCSGVTFQS
AVKRTIKGTHHWLLLTILTSLLVLVQSTQWSLFFFLYENAFLPFAMGIIAMSAFAMMFVKHKHAFLCLFL
LPSLATVAYFNMVYMPASWVMRIMTWLDMVDTSLSGFKLKDCVMYASAVVLLILMTARTVYDDGARRVWT
LMNVLTLVYKVYYGNALDQAISMWALIISVTSNYSGVVTTVMFLARGIVFMCVEYCPIFFITGNTLQCIM
LVYCFLGYFCTCYFGLFCLLNRYFRLTLGVYDYLVSTQEFRYMNSQGLLPPKNSIDAFKLNIKLLGVGGK
PCIKVATVQSKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLLS
MQGAVDINKLCEEMLDNRATLQAIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSE
FDRDAAMQRKLEKMADQAMTQMYKQARSEDKRAKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLN
IIPLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVVDADSKIVQLSEISMDNSPNLAWPLIVTA
LRANSAVKLQNNELSPVALRQMSCAAGTTQTACTDDNALAYYNTTKGGRFVLALLSDLQDLKWARFPKSD
GTGTIYTELEPPCRFVTDTPKGPKVKYLYFIKGLNNLNRGMVLGSLAATVRLQAGNATEVPANSTVLSFC
AFAVDAAKAYKDYLASGGQPITNCVKMLCTHTGTGQAITVTPEANMDQESFGGASCCLYCRCHIDHPNPK
GFCDLKGKYVQIPTTCANDPVGFTLKNTVCTVCGMWKGYGCSCDQLREPMLQSADAQSFLNGFAV"""
Show/Hide Solution
Given the string “nOBody Said iT was eAsy, No oNe Ever saId it WoulD be tHis hArd…”
Create a list with all the letters that are capitalized (use str.isupper)
Print the list
Use the string method join to concatenate all the letters in a string, using “*” as separator. The syntax of join is str.join(list) and it outputs a string with all the elements in list joined with the character in str (es. “+”.join([1,2,3]) returns “1+2+3”).
The expected output:
['O', 'B', 'S', 'T', 'A', 'N', 'N', 'E', 'I', 'W', 'D', 'H', 'A']
O*B*S*T*A*N*N*E*I*W*D*H*A
Show/Hide Solution
Given the following sequence:
AUGCUGUCUCCCUCACUGUAUGUAAAUUGCAUCUAGAAUAGCA
UCUGGAGCACUAAUUGACACAUAGUGGGUAUCAAUUAUUA
UUCCAGGUACUAGAGAUACCUGGACCAUUAACGGAUAAAU
AGAAGAUUCAUUUGUUGAGUGACUGAGGAUGGCAGUUCCU
GCUACCUUCAAGGAUCUGGAUGAUGGGGAGAAACAGAGAA
CAUAGUGUGAGAAUACUGUGGUAAGGAAAGUACAGAGGAC
UGGUAGAGUGUCUAACCUAGAUUUGGAGAAGGACCUAGAA
GUCUAUCCCAGGGAAAUAAAAAUCUAAGCUAAGGUUUGAG
GAAUCAGUAGGAAUUGGCAAAGGAAGGACAUGUUCCAGAU
GAUAGGAACAGGUUAUGCAAAGAUCCUGAAAUGGUCAGAG
CUUGGUGCUUUUUGAGAACCAAAAGUAGAUUGUUAUGGAC
CAGUGCUACUCCCUGCCUCUUGCCAAGGGACCCCGCCAAG
CACUGCAUCCCUUCCCUCUGACUCCACCUUUCCACUUGCC
CAGUAUUGUUGGUG
Show/Hide Solution
Given the following list of gene correlations:
geneCorr = [["G1C2W9", "G1C2Q7", 0.2], ["G1C2W9", "G1C2Q4", 0.9], ["Q6NMS1", "G1C2W9", 0.8],["G1C2W9", "Q6NMS1",0.4], ["G1C2Q7", "G1C2Q4",0.76]]
where each sublist [“gene1”, “gene2”, corr] represents a correlation between gene1 and gene2 with correlation corr, create another list containing only the elements having an high correlation (i.e. > 0.75). Print this list.
Expected result:
[['G1C2W9', 'G1C2Q4', 0.9], ['Q6NMS1', 'G1C2W9', 0.8], ['G1C2Q7', 'G1C2Q4', 0.76]]
Show/Hide Solution
Given the following sequence of DNA:
DNA = “GATTACATATATCAGTACAGATATATACGCGCGGGCTTACTATTAAAAACCCC”
1. Create a dictionary reporting the frequency of each base (i.e. key is the
base and value is the frequency).
2. Create a dictionary representing an index of all possible dimers (i.e. 2
bases, 16 dimers in total): AA, AT, AC, AG, TA, TT, TC, TG, ... . In this case,
keys of the dictionary are dimers and values are lists with all possible starting
positions of the dimer.
3. Print the DNA string.
4. Print for each base its frequency
4. Print all positions of the dimer "AT"
The expected result is:
sequence: GATTACATATATCAGTACAGATATATACGCGCGGGCTTACTATTAAAAACCCC
G has frequency: 0.1509433962264151
C has frequency: 0.22641509433962265
A has frequency: 0.3584905660377358
T has frequency: 0.2641509433962264
{'GG': [32, 33], 'TC': [11], 'GT': [14], 'CA': [5, 12, 17], 'TT': [2, 36, 42],
'CG': [27, 29, 31], 'TA': [3, 7, 9, 15, 21, 23, 25, 37, 40, 43], 'AG': [13, 18],
'GA': [0, 19], 'CT': [35, 39], 'GC': [28, 30, 34], 'AT': [1, 6, 8, 10, 20, 22, 24, 41],
'CC': [49, 50, 51], 'AA': [44, 45, 46, 47], 'AC': [4, 16, 26, 38, 48]}
Dimer AT is found at: [1, 6, 8, 10, 20, 22, 24, 41]
Show/Hide Solution
Given the following table, reporting molecular weights for each amino acid, store them in a dictionary where the key is the one letter code and the value is the molecular weight (e.g. {“A” : 89, “R”:174”}).
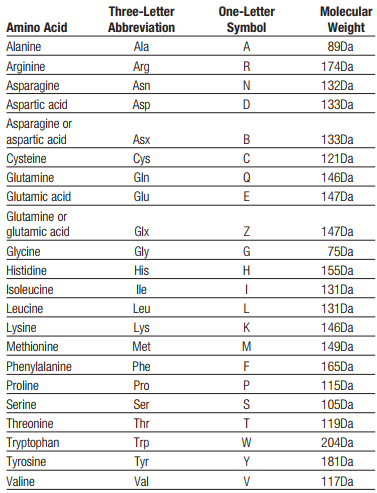
Write a python script to answer the following questions:
1. What is the average molecular weight of an amino acid?
2. What is the total molecular weight and number of aminoacids
of the P53 peptide GSRAHSSHLKSKKGQSTSRHK?
3. What is the total molecular weight and number of aminoacids
of the peptide YTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWF?
Show/Hide Solution
The following string is an extract of a blast alignment with compacted textual output. This is a comma (“,”) separated text file where the columns report the following info: the first column is the query_id, the second the subject_id (i.e. the reference on which we aligned the query), the third is the percentage of identity and then we have the alignment length, number of mismatches, gap opens, start point of the alignment on the query, end point of the alignment on the query, start point of the alignment on the subject, end point of the alignment on the subject and evalue of the alignment.
#Fields:q.id,s.id,% ident,align len,mismatches,gap opens,q.start,q.end,s.start,s.end,evalue
ab1_400a,scaffold16155,98.698,384,4,1,12,394,6700,7083,0.0
ab1_400b,scaffold14620,98.698,384,4,1,12,394,1240,857,0.0
92A2_SP6_344a,scaffold14394,95.575,113,5,0,97,209,250760,250648,2.92e-44
92A2_SP6_344b,scaffold10682,97.849,93,2,0,18,110,898,990,3.81e-38
92A2_T7_558a,scaffold277,88.746,311,31,3,21,330,26630,26937,5.81e-103
92A2_T7_558b,scaffold277,89.545,220,21,2,27,246,27167,26950,6.06e-73
92A2_T7_558c,scaffold1125,88.125,320,31,5,30,346,231532,231847,7.51e-102
ab1_675a,scaffold4896,100.000,661,0,0,15,675,79051,78391,0.0
ab1_676b,scaffold4896,99.552,670,0,3,7,673,78421,79090,0.0
For each alignment, store the subject id, the percentage of identity and evalue, subject start and end in a dictionary using the query id as key. All this information can be stored in a dictionary having subject id as key and a dictionary with all the information as value:
alignments["ab1_400"] = {"subjectid" : scaffold16155, "perc_id" : 98.698, "evalue" : 0.0}Print the whole dictionary
Print only the alignments having percentage of identity > 90%
Note: skip the first comment line (i.e. skip line if starts with “#”). Note1: when storing the percentage of identity remember to convert the string into a float.
The expected output is:
{'92A2_T7_558a': {'perc_id': 88.746, 'subjectid': 'scaffold277', 'evalue': '5.81e-103'},
'92A2_T7_558b': {'perc_id': 89.545, 'subjectid': 'scaffold277', 'evalue': '6.06e-73'},
'92A2_SP6_344b': {'perc_id': 97.849, 'subjectid': 'scaffold10682', 'evalue': '3.81e-38'},
'92A2_T7_558c': {'perc_id': 88.125, 'subjectid': 'scaffold1125', 'evalue': '7.51e-102'},
'ab1_400a': {'perc_id': 98.698, 'subjectid': 'scaffold16155', 'evalue': '0.0'},
'ab1_675a': {'perc_id': 100.0, 'subjectid': 'scaffold4896', 'evalue': '0.0'},
'ab1_400b': {'perc_id': 98.698, 'subjectid': 'scaffold14620', 'evalue': '0.0'},
'92A2_SP6_344a': {'perc_id': 95.575, 'subjectid': 'scaffold14394', 'evalue': '2.92e-44'},
'ab1_676b': {'perc_id': 99.552, 'subjectid': 'scaffold4896', 'evalue': '0.0'}}
Alignments with identity > 90%:
Query id Subject id % ident evalue
92A2_SP6_344b scaffold10682 97.849 3.81e-38
ab1_400a scaffold16155 98.698 0.0
ab1_675a scaffold4896 100.0 0.0
ab1_400b scaffold14620 98.698 0.0
92A2_SP6_344a scaffold14394 95.575 2.92e-44
ab1_676b scaffold4896 99.552 0.0
Show/Hide Solution
The following text (separated with a comma “,”) is an extract of protein-protein interactions network stored in the database STRING involving PKLR (Pyruvate kinase, liver and RBC) that plays a key role in glycolysis:
#node1,node2,node1_ext_id,node2_ext_id,
ENO1,TPI1,ENSP00000234590,ENSP00000229270
PKLR,ENO1,ENSP00000339933,ENSP00000234590
PKLR,ENO3,ENSP00000339933,ENSP00000324105
PGK1,ENO1,ENSP00000362413,ENSP00000234590
PGK1,TPI1,ENSP00000362413,ENSP00000229270
GPI,TPI1,ENSP00000405573,ENSP00000229270
PKLR,ENO2,ENSP00000339933,ENSP00000229277
PGK1,ENO3,ENSP00000362413,ENSP00000324105
PGK1,ENO2,ENSP00000362413,ENSP00000229277
GPI,PKLR,ENSP00000405573,ENSP00000339933
ENO2,TPI1,ENSP00000229277,ENSP00000229270
PGK2,ENO1,ENSP00000305995,ENSP00000234590
ENO3,PGK2,ENSP00000324105,ENSP00000305995
PGK2,TPI1,ENSP00000305995,ENSP00000229270
ENO3,TPI1,ENSP00000324105,ENSP00000229270
PGK2,ENO2,ENSP00000305995,ENSP00000229277
GPI,ENO3,ENSP00000405573,ENSP00000324105
PKLR,LDHB,ENSP00000339933,ENSP00000229319
PKLR,LDHC,ENSP00000339933,ENSP00000280704
PKLR,TPI1,ENSP00000339933,ENSP00000229270
PGK1,PKLR,ENSP00000362413,ENSP00000339933
GPI,ENO2,ENSP00000405573,ENSP00000229277
PKLR,PGK2,ENSP00000339933,ENSP00000305995
GPI,PGK1,ENSP00000405573,ENSP00000362413
ME3,PKLR,ENSP00000352657,ENSP00000339933
ME3,LDHB,ENSP00000352657,ENSP00000229319
ME3,LDHC,ENSP00000352657,ENSP00000280704
GPI,PGK2,ENSP00000405573,ENSP00000305995
GPI,ENO1,ENSP00000405573,ENSP00000234590
GPI,LDHB,ENSP00000405573,ENSP00000229319
ENO3,ENO2,ENSP00000324105,ENSP00000229277
GPI,LDHC,ENSP00000405573,ENSP00000280704
ENO3,LDHB,ENSP00000324105,ENSP00000229319
ENO3,LDHC,ENSP00000324105,ENSP00000280704
ENO1,LDHB,ENSP00000234590,ENSP00000229319
LDHB,TPI1,ENSP00000229319,ENSP00000229270
LDHC,TPI1,ENSP00000280704,ENSP00000229270
PGK2,LDHC,ENSP00000305995,ENSP00000280704
PGK1,LDHB,ENSP00000362413,ENSP00000229319
PGK1,PGK2,ENSP00000362413,ENSP00000305995
ENO1,ENO2,ENSP00000234590,ENSP00000229277
LDHC,ENO1,ENSP00000280704,ENSP00000234590
LDHB,ENO2,ENSP00000229319,ENSP00000229277
LDHC,ENO2,ENSP00000280704,ENSP00000229277
ENO3,ENO1,ENSP00000324105,ENSP00000234590
PGK1,LDHC,ENSP00000362413,ENSP00000280704
GPI,ME3,ENSP00000405573,ENSP00000352657
PGK2,LDHB,ENSP00000305995,ENSP00000229319
ME3,TPI1,ENSP00000352657,ENSP00000229270
Here is a graphic representation of the protein-protein interactions:
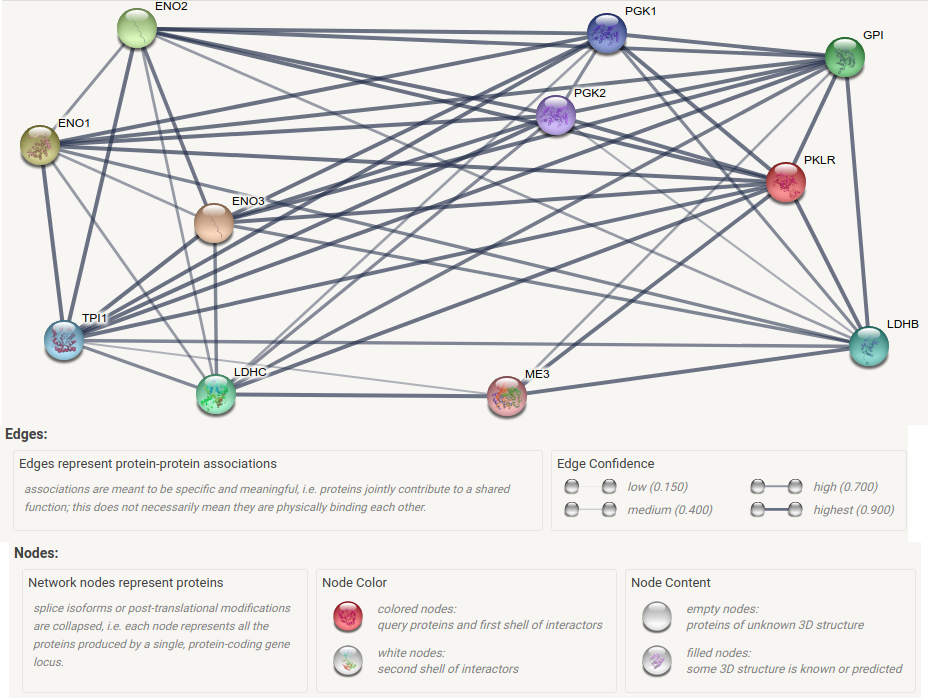
Note: we can assume that relations between nodes are transitive. node1 --> node2
implies node2 --> node1.
1. Store the network information in a dictionary having node1 as key and
the list of all nodes2 associated to it as value (remember to skip the first
line that is the header). Remember transitivity, therefore add also node2 -->node1
2. Find all first neighbours of "PKLR" (i.e. the nodes that are directly connected
to "PKLR") and print them
3. Find all first neighbours of "ME3" (i.e. the nodes that are directly connected
to "PKLR") and print them
4. Find all the second neighbours of "ME3" (i.e. the nodes that are connected to nodes
directly connected to "ME3", but not directly to "ME3").
Show/Hide Solution